Why can't we be friends? Plotting frequentist (lmerTest) and Bayesian (brms) mixed-effects models
Frequentist and Bayesian statistics are sometimes regarded as fundamentally different philosophies. Indeed, can both methods qualify as philosophies, or is one of them just a pointless ritual? Is frequentist statistics about lmerTest and brms are simultaneously loaded? If only we could fit frequentist and Bayesian models to the same data and plot the results together, we might get a glimpse into these puzzles.
All the analyses shown below can be reproduced using the materials at https://osf.io/gt5uf. The combination of the frequentist and the Bayesian estimates in the same plot is achieved using the following custom function from Bernabeu (2022).
Visualising frequentist and Bayesian estimates in one plot
Both frequentist and Bayesian statistics offer the options of hypothesis testing and parameter estimation (Cumming, 2014; Kruschke & Liddell, 2018; Rouder et al., 2018; Schmalz et al., 2022; Tendeiro & Kiers, 2019, 2022; van Ravenzwaaij & Wagenmakers, 2022). In the statistical analyses conducted by Bernabeu (2022), hypothesis testing was performed within the frequentist framework, whereas parameter estimation was performed within both the frequentist and the Bayesian frameworks (for other examples of the estimation approach, see Milek et al., 2018; Pregla et al., 2021; Rodríguez-Ferreiro et al., 2020).
# Presenting frequentist and Bayesian estimates in the same plot. For this # purpose, the frequentist results are merged into a plot from # brms::mcmc_plot(), Three arguments are required for this function: a # summary of an 'lmerTest' model, confidence intervals from # lme4::confint.merMod(), and a plot from brms::mcmc_plot(). The function # is equipped to accept the slight differences between the names of the # predictors in the frequentist results and in the Bayesian results. frequentist_bayesian_plot = function( frequentist_model_summary, confidence_intervals, mcmc_plot, labels = NULL, font_size = 10, x_title = 'Estimate', x_axis_labels = 6, # <-- approximate number of labels in X axis # Number of columns into which the legend labels (i.e., # 'Frequentist analysis', 'Bayesian analysis') should # be distributed. legend_ncol = 2, # If note_frequentist_no_prior = TRUE, '(no prior)' is appended to the # label 'Frequentist analysis' in the legend. This is useful when # the plot is titled with the prior, to clarify that the prior # does not concern the frequentist analysis. note_frequentist_no_prior = FALSE, # If interaction_symbol_x = TRUE, replace double colons with times # symbols followed by line breaks and indentation. Then, replace # single colons with times symbols. interaction_symbol_x = FALSE, # Vertical line at specified value of X axis vertical_line_at_x = NULL ) { require(dplyr) require(forcats) require(ggplot2) require(stringr) require(ggtext) # Format frequentist results to match the format of mcmc_plot # Firstly, remove parentheses from the intercept and add 'b_' # to the beginning of every predictor rownames(frequentist_model_summary$coefficients) = rownames(frequentist_model_summary$coefficients) %>% str_replace(pattern = '\\(Intercept\\)', replacement = 'Intercept') %>% str_replace(pattern = '^', replacement = 'b_') rownames(confidence_intervals) = rownames(confidence_intervals) %>% str_replace(pattern = '\\(Intercept\\)', replacement = 'Intercept') %>% str_replace(pattern = '^', replacement = 'b_') # Next, create 'parameters' column in the frequentist results frequentist_model_summary$coefficients = frequentist_model_summary$coefficients %>% as.data.frame() %>% mutate(parameter = rownames(frequentist_model_summary$coefficients)) %>% rename(frequentist_estimate = Estimate) %>% data.frame(row.names = NULL) confidence_intervals = confidence_intervals %>% as.data.frame() %>% mutate(parameter = rownames(confidence_intervals)) %>% rename(CI_2.5 = '2.5 %', CI_97.5 = '97.5 %') %>% data.frame(row.names = NULL) # Merge both frequentist objects into mcmc_plot mcmc_plot$data = list(mcmc_plot$data, frequentist_model_summary$coefficients %>% select(parameter, frequentist_estimate), confidence_intervals) %>% purrr::reduce(full_join, by = 'parameter') # If labels supplied and interaction_symbol_x = TRUE, replace double # colons with times symbols followed by line breaks and indentation. # Then, replace single colons with times symbols. if(!is.null(labels) & isTRUE(interaction_symbol_x)) { labels = labels %>% gsub('::', ' × <br> ', .) %>% gsub(':', ' × ', .) } # Set current order of effects to prevent alphabetical order labels = factor(labels, levels = labels) # Plot plot = mcmc_plot + # Add vertical line (on top of default line from mcmc_plot) geom_vline(xintercept = vertical_line_at_x, col = 'grey60') + # Add confidence intervals from frequentist model geom_errorbarh(aes(xmin = CI_2.5, xmax = CI_97.5), height = 0.12, position = position_nudge(y = 0.09), colour = '#ff7474ff') + # Add frequentist estimate geom_point(aes(x = frequentist_estimate), position = position_nudge(y = 0.09), colour = 'red', size = 0.8) + scale_x_continuous(n.breaks = x_axis_labels, limits = c(min(mcmc_plot$data$CI_2.5, mcmc_plot$data$x), max(mcmc_plot$data$CI_97.5, mcmc_plot$data$x))) + scale_y_discrete(expand = c(0.023, 0.033)) + # X-axis title xlab(x_title) + theme_minimal() + theme(text = element_text(family = 'Arial'), # <-- prevent R crashing (see https://discourse.mc-stan.org/t/error-code-when-using-pp-check-with-brms/27419/4) plot.title = element_markdown(colour = '#005b96', hjust = 0.5, vjust = 1, size = font_size * 1.2), axis.title.x = ggtext::element_markdown(size = font_size, margin = margin(t = font_size * 0.8)), axis.text.x = element_text(size = font_size * 0.9, margin = margin(t = font_size * 0.4)), axis.title.y = element_blank(), # Use 'vjust' used to place each Y axis label at the foot of its respective segment axis.text.y = ggtext::element_markdown(size = font_size, vjust = 0), axis.ticks.x = element_line(), # Parameters are adjusted based on number of columns in legend using ifelse() legend.text = element_text(size = font_size, lineheight = ifelse(legend_ncol == 1, 0.3, 1.2), margin = margin(r = ifelse(legend_ncol == 1, 2, 4))), legend.key.height = unit(ifelse(legend_ncol == 1, 15, 21), 'pt'), legend.background = element_rect(colour = 'grey70', fill = 'white'), panel.grid.major.y = element_line(colour = 'grey90'), plot.margin = margin(9, 4, 14, 12)) # Manual legend for frequentist analysis (red) and Bayesian analysis (blue), # subject to the presence of 'note_frequentist_no_prior'. if(note_frequentist_no_prior) { plot = plot + geom_line(aes(colour = factor(parameter)), size = 0) + # <-- placeholder to allow manual scale scale_colour_manual(values = c('Frequentist analysis\n(no prior)' = 'red', 'Bayesian analysis' = '#005b96'), # <-- fancy blue guide = guide_legend(title = NULL, override.aes = list(size = 5), ncol = legend_ncol)) } else{ plot = plot + geom_line(aes(colour = factor(parameter)), size = 0) + # <-- placeholder to allow manual scale scale_colour_manual(values = c('Frequentist analysis' = 'red', 'Bayesian analysis' = '#005b96'), # <-- fancy blue guide = guide_legend(title = NULL, override.aes = list(size = 4), ncol = legend_ncol)) } # If labels supplied, pass them to 'scale_y_discrete'. This is necessary # because modifying the 'parameter' column directly creates errors in # the plot. if(!is.null(labels)) { plot = plot + # Apply modified labels, in a reverse order to match correct order pre-established # by the 'mcmc_plot' function that was used to create the original plots # (see https://discourse.mc-stan.org/t/manually-changing-y-axis-tick-labels-in-brms-mcmc-plot/19727/5) scale_y_discrete(labels = rev(labels), limits = rev, # Padding at the borders expand = c(0.023, 0.033)) } # Output plot }
Let’s load the function from GitHub and put it to the test.
Hide
# Presenting the frequentist and the Bayesian estimates in the same plot.
# For this purpose, the frequentist results are merged into a plot from
# brms::mcmc_plot()
source('https://raw.githubusercontent.com/pablobernabeu/language-sensorimotor-simulation-PhD-thesis/main/R_functions/frequentist_bayesian_plot.R')
# install.packages('devtools')
# library(devtools)
# install_version('tidyverse', '1.3.1') # Due to breaking changes, Version 1.3.1 is required.
# install_version('ggplot2', '3.3.5') # Due to breaking changes, Version 3.3.5 is required.
library(tidyverse)
library(ggplot2)
library(Cairo)
# Load frequentist coefficients (estimates and confidence intervals)
KR_summary_semanticpriming_lmerTest =
readRDS(gzcon(url('https://github.com/pablobernabeu/language-sensorimotor-simulation-PhD-thesis/blob/main/semanticpriming/frequentist_analysis/results/KR_summary_semanticpriming_lmerTest.rds?raw=true')))
confint_semanticpriming_lmerTest =
readRDS(gzcon(url('https://github.com/pablobernabeu/language-sensorimotor-simulation-PhD-thesis/blob/main/semanticpriming/frequentist_analysis/results/confint_semanticpriming_lmerTest.rds?raw=true')))
# Below are the default names of the effects
# rownames(KR_summary_semanticpriming_lmerTest$coefficients)
# rownames(confint_semanticpriming_lmerTest)
# Load Bayesian posterior distributions
semanticpriming_posteriordistributions_weaklyinformativepriors_exgaussian =
readRDS(gzcon(url('https://github.com/pablobernabeu/language-sensorimotor-simulation-PhD-thesis/blob/main/semanticpriming/bayesian_analysis/results/semanticpriming_posteriordistributions_weaklyinformativepriors_exgaussian.rds?raw=true')))
# Below are the default names of the effects
# levels(semanticpriming_posteriordistributions_weaklyinformativepriors_exgaussian$data$parameter)
# Reorder the components of interactions in the frequentist results to match
# with the order present in the Bayesian results.
rownames(KR_summary_semanticpriming_lmerTest$coefficients) =
rownames(KR_summary_semanticpriming_lmerTest$coefficients) %>%
str_replace(pattern = 'z_recoded_interstimulus_interval:z_cosine_similarity',
replacement = 'z_cosine_similarity:z_recoded_interstimulus_interval') %>%
str_replace(pattern = 'z_recoded_interstimulus_interval:z_visual_rating_diff',
replacement = 'z_visual_rating_diff:z_recoded_interstimulus_interval')
rownames(confint_semanticpriming_lmerTest) =
rownames(confint_semanticpriming_lmerTest) %>%
str_replace(pattern = 'z_recoded_interstimulus_interval:z_cosine_similarity',
replacement = 'z_cosine_similarity:z_recoded_interstimulus_interval') %>%
str_replace(pattern = 'z_recoded_interstimulus_interval:z_visual_rating_diff',
replacement = 'z_visual_rating_diff:z_recoded_interstimulus_interval')
# Create a vector containing the names of the effects. This vector will be passed
# to the plotting function.
new_labels =
semanticpriming_posteriordistributions_weaklyinformativepriors_exgaussian$data$parameter %>%
unique %>%
# Remove the default 'b_' from the beginning of each effect
str_remove('^b_') %>%
# Put Intercept in parentheses
str_replace(pattern = 'Intercept', replacement = '(Intercept)') %>%
# First, adjust names of variables (both in main effects and in interactions)
str_replace(pattern = 'z_target_word_frequency',
replacement = 'Target-word frequency') %>%
str_replace(pattern = 'z_target_number_syllables',
replacement = 'Number of target-word syllables') %>%
str_replace(pattern = 'z_word_concreteness_diff',
replacement = 'Word-concreteness difference') %>%
str_replace(pattern = 'z_cosine_similarity',
replacement = 'Language-based similarity') %>%
str_replace(pattern = 'z_visual_rating_diff',
replacement = 'Visual-strength difference') %>%
str_replace(pattern = 'z_attentional_control',
replacement = 'Attentional control') %>%
str_replace(pattern = 'z_vocabulary_size',
replacement = 'Vocabulary size') %>%
str_replace(pattern = 'z_recoded_participant_gender',
replacement = 'Gender') %>%
str_replace(pattern = 'z_recoded_interstimulus_interval',
replacement = 'SOA') %>%
# Show acronym in main effect of SOA
str_replace(pattern = '^SOA$',
replacement = 'Stimulus onset asynchrony (SOA)') %>%
# Second, adjust order of effects in interactions. In the output from the model,
# the word-level variables of interest (i.e., 'z_cosine_similarity' and
# 'z_visual_rating_diff') sometimes appeared second in their interactions. For
# better consistency, the code below moves those word-level variables (with
# their new names) to the first position in their interactions. Note that the
# order does not affect the results in any way.
sub('(\\w+.*):(Language-based similarity|Visual-strength difference)',
'\\2:\\1',
.) %>%
# Replace colons denoting interactions with times symbols
str_replace(pattern = ':', replacement = ' × ')
# Create plot
plot_semanticpriming_frequentist_bayesian_plot_weaklyinformativepriors_exgaussian =
frequentist_bayesian_plot(KR_summary_semanticpriming_lmerTest,
confint_semanticpriming_lmerTest,
semanticpriming_posteriordistributions_weaklyinformativepriors_exgaussian,
labels = new_labels, interaction_symbol_x = TRUE,
vertical_line_at_x = 0, x_title = 'Effect size (β)',
legend_ncol = 1) +
theme(legend.position = 'bottom')Hide
plot_semanticpriming_frequentist_bayesian_plot_weaklyinformativepriors_exgaussian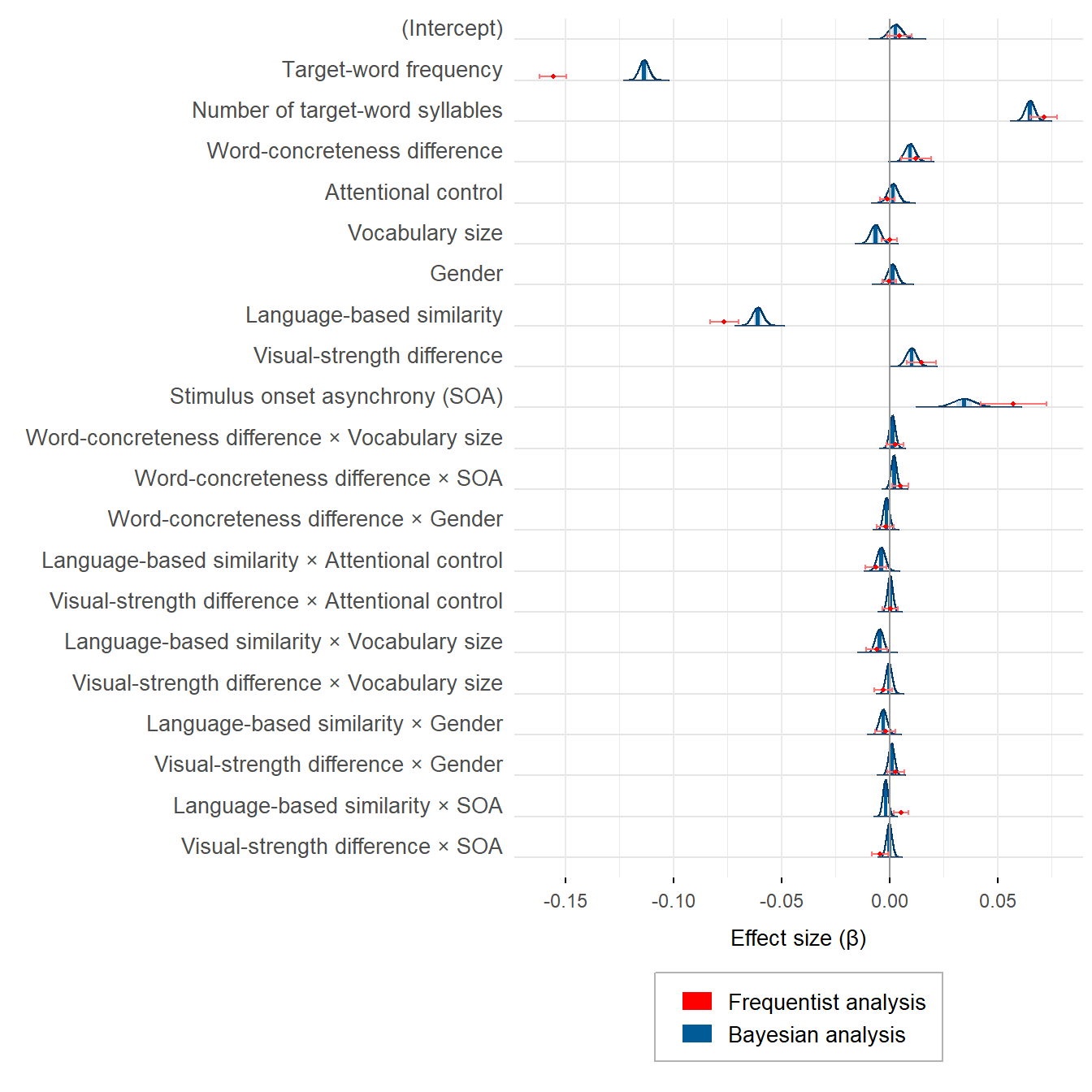
Frequentist and Bayesian estimates are not so polar opposites, are they? What is more, the larger differences between some estimates are the result of the priors that were set on the corresponding effects. With uninformative priors, the frequentist and the Bayesian estimates are virtually identical.
# Define priors # In the function `set_prior` used below, the argument `class` often represents # groups of parameters. For instance, class 'b' refers to fixed effects. # Since most priors do not specify the negative/positive direction of effects, # distributions are first specified for entire classes, and those are # overridden wherever a direction is specified. weaklyinformative_priors = c(set_prior('normal(0, 0.2)', class = 'Intercept'), set_prior('normal(0, 0.2)', class = 'b'), set_prior('normal(0.1, 0.2)', class = 'b', coef = 'z_target_number_syllables'), set_prior('normal(-0.1, 0.2)', class = 'b', coef = 'z_target_word_frequency'), set_prior('normal(0, 0.2)', class = 'sd'), # automatically truncated to keep positive values only set_prior('normal(0, 0.2)', class = 'sigma'), # automatically truncated to keep positive values only set_prior('lkj(2)', class = 'cor') # standard, regularising prior on random-effects covariance to aid convergence )
Now it’s time to consider in earnest:
Is frequentist statistics about
values only? Are frequentist estimates diametrically opposed to Bayesian posterior distributions? Are confidence intervals and credible intervals irreconcilable? Will R crash if lmerTestandbrmsare simultaneously loaded?
Session info
If you encounter any blockers while reproducing the above analyses using the materials at https://osf.io/gt5uf, my current session info may be useful. For instance, the legend of the plot may not show if the latest versions of the ggplot2 and the tidyverse packages are used. Instead, ggplot2 3.3.5 and tidyverse 1.3.1 should be installed using install_version('ggplot2', '3.3.5') and install_version('tidyverse', '1.3.1').
Hide
sessionInfo()## R version 4.2.3 (2023-03-15 ucrt)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows 10 x64 (build 22621)
##
## Matrix products: default
##
## locale:
## [1] LC_COLLATE=English_United Kingdom.utf8
## [2] LC_CTYPE=English_United Kingdom.utf8
## [3] LC_MONETARY=English_United Kingdom.utf8
## [4] LC_NUMERIC=C
## [5] LC_TIME=English_United Kingdom.utf8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] ggtext_0.1.2 Cairo_1.6-0 forcats_1.0.0
## [4] stringr_1.5.0 dplyr_1.1.1 purrr_1.0.1
## [7] readr_2.1.4 tidyr_1.3.0 tibble_3.2.1
## [10] ggplot2_3.3.5 tidyverse_1.3.1 knitr_1.42
## [13] xaringanExtra_0.7.0
##
## loaded via a namespace (and not attached):
## [1] Rcpp_1.0.10 lubridate_1.9.2 digest_0.6.31 utf8_1.2.3
## [5] plyr_1.8.8 R6_2.5.1 cellranger_1.1.0 ggridges_0.5.4
## [9] backports_1.4.1 reprex_2.0.2 evaluate_0.21 highr_0.10
## [13] httr_1.4.6 blogdown_1.16 pillar_1.9.0 rlang_1.1.0
## [17] uuid_1.1-0 readxl_1.4.2 rstudioapi_0.14 jquerylib_0.1.4
## [21] rmarkdown_2.21 labeling_0.4.2 munsell_0.5.0 gridtext_0.1.5
## [25] broom_1.0.4 compiler_4.2.3 modelr_0.1.11 xfun_0.38
## [29] pkgconfig_2.0.3 htmltools_0.5.5 tidyselect_1.2.0 bookdown_0.33.3
## [33] fansi_1.0.4 crayon_1.5.2 tzdb_0.4.0 dbplyr_2.3.2
## [37] withr_2.5.0 commonmark_1.9.0 grid_4.2.3 jsonlite_1.8.4
## [41] gtable_0.3.3 lifecycle_1.0.3 DBI_1.1.3 magrittr_2.0.3
## [45] scales_1.2.1 cli_3.4.1 stringi_1.7.12 cachem_1.0.7
## [49] farver_2.1.1 fs_1.6.1 xml2_1.3.3 bslib_0.4.2
## [53] generics_0.1.3 vctrs_0.6.1 tools_4.2.3 glue_1.6.2
## [57] markdown_1.5 hms_1.1.3 fastmap_1.1.1 yaml_2.3.7
## [61] timechange_0.2.0 colorspace_2.1-0 rvest_1.0.3 haven_2.5.2
## [65] sass_0.4.6References
Bernabeu, P. (2022). Language and sensorimotor simulation in conceptual processing: Multilevel analysis and statistical power. Lancaster University. https://doi.org/10.17635/lancaster/thesis/1795
Cumming, G. (2014). The new statistics: Why and how. Psychological Science, 25(1), 7–29. https://doi.org/10.1177/0956797613504966
Kruschke, J. K., & Liddell, T. M. (2018). The Bayesian New Statistics: Hypothesis testing, estimation, meta-analysis, and power analysis from a Bayesian perspective. Psychonomic Bulletin & Review, 25(1), 178–206.
Milek, A., Butler, E. A., Tackman, A. M., Kaplan, D. M., Raison, C. L., Sbarra, D. A., Vazire, S., & Mehl, M. R. (2018). “Eavesdropping on happiness” revisited: A pooled, multisample replication of the association between life satisfaction and observed daily conversation quantity and quality. Psychological Science, 29(9), 1451–1462. https://doi.org/10.1177/0956797618774252
Pregla, D., Lissón, P., Vasishth, S., Burchert, F., & Stadie, N. (2021). Variability in sentence comprehension in aphasia in German. Brain and Language, 222, 105008. https://doi.org/10.1016/j.bandl.2021.105008
Rodríguez-Ferreiro, J., Aguilera, M., & Davies, R. (2020). Semantic priming and schizotypal personality: Reassessing the link between thought disorder and enhanced spreading of semantic activation. PeerJ, 8, e9511. https://doi.org/10.7717/peerj.9511
Rouder, J. N., Haaf, J. M., & Vandekerckhove, J. (2018). Bayesian inference for psychology, part IV: Parameter estimation and Bayes factors. Psychonomic Bulletin & Review, 25(1), 102–113. https://doi.org/10.3758/s13423-017-1420-7
Schmalz, X., Biurrun Manresa, J., & Zhang, L. (2021). What is a Bayes factor? Psychological Methods. https://doi.org/10.1037/met0000421
Tendeiro, J. N., & Kiers, H. A. L. (2019). A review of issues about null hypothesis Bayesian testing. Psychological Methods, 24(6), 774–795. https://doi.org/10.1037/met0000221
Tendeiro, J. N., & Kiers, H. A. L. (2022). On the white, the black, and the many shades of gray in between: Our reply to van Ravenzwaaij and Wagenmakers (2021). Psychological Methods, 27(3), 466–475. https://doi.org/10.1037/met0000505
van Ravenzwaaij, D., & Wagenmakers, E.-J. (2022). Advantages masquerading as “issues” in Bayesian hypothesis testing: A commentary on Tendeiro and Kiers (2019). Psychological Methods, 27(3), 451–465. https://doi.org/10.1037/met0000415